Generative biology: new approaches to study developmental design principles
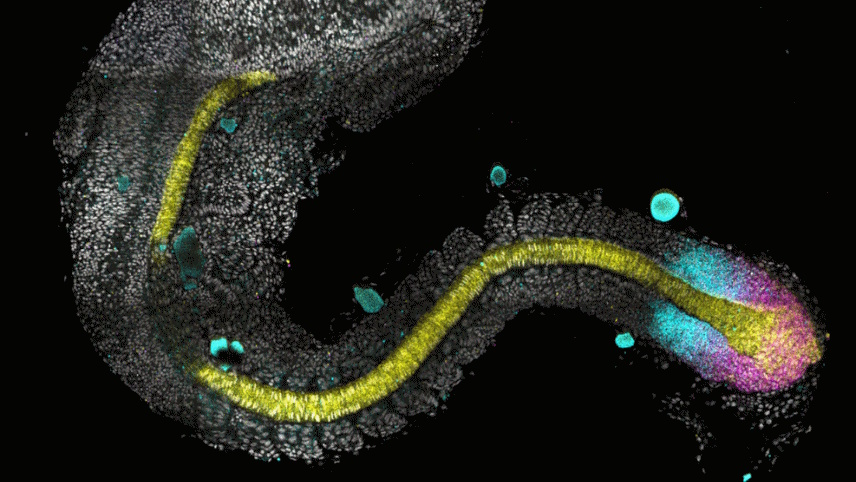
Theo Murphy meeting organised by Dr Jake Cornwall-Scoones, Dr Dirk Benzinger, and Professor Sally Lowell.
As systems-level measurement of embryogenesis reaches maturity, developmental biologists are returning to foundational questions of how embryos build themselves. Synthetic biology has demonstrated how bottom-up explanations reveal design principles of biological transitions, and is increasingly looking towards embryos for inspiration, holistic contextualisation, and evolutionary interpretation. This meeting brings together these two disciplines towards a science of generative biology.
Programme
The programme, including speaker biographies and abstracts, will be available soon. Please note the programme may be subject to change.
Poster session
There will be a poster session on Monday 20 October. If you would like to present a poster, please submit your proposed title, abstract (up to 200 words), author list, and the name of the proposed presenter and institution to the Scientific Programmes team. Acceptances may be made on a rolling basis so we recommend submitting as soon as possible in case the session becomes full. Submissions made within one month of the meeting may not be included in the programme booklet.
Short talks
The meeting organisers are also accepting proposals for 10 minute talks to be delivered as part of the meeting programme. If you would like to present a talk, please submit your proposed title and abstract (up to 200 words) to the Scientific Programmes team. The deadline for submissions is Friday 15 August.
Attending this event
- Free to attend and in-person only
- When requesting an invitation, please briefly state your expertise and reasons for attending
- Requests are reviewed by the meeting organisers on a rolling basis. You will receive a link to register if your request has been successful
- Catering options will be available to purchase upon registering. Participants are responsible for booking their own accommodation. Please do not book accommodation until you have been invited to attend the meeting by the meeting organisers
Enquiries: contact the Scientific Programmes team.
Image credit:©️Shannon Taylor / Oxford, Verd Lab
Organisers
Schedule
| 09:00-09:05 |
Welcome by the organisers
|
|---|---|
| 09:05-09:25 |
Mechanical regulation of cell states
The structure of tissues is tightly linked to their function. During formation of functional organs, large-scale changes in tissue elongation, stretching, compression, folding/buckling, and budding impact the shape, position, packing, and contractility state of cells. Conversely, changes in single cell contractility, shape and position locally alter tissue organization and mechanics. Thus, forces function as important ques that are transmitted to the nucleus to coordinate gene expression programs to control cell states. On the other hand, excessive mechanical stresses have the potential to damage cells and tissues. In my presentation I will discuss our recent research on how cells sense mechanical forces and how these mechanosignals are integrated with biochemical inputs to alter cell states in health and disease. Dr Sara WickströmMax Planck Institute for Molecular Biomedicine, Germany Dr Sara WickströmMax Planck Institute for Molecular Biomedicine, Germany Sara Wickström received her MD in 2001 and PhD in 2004 from the University of Helsinki, Finland. After postdoctoral training at the Max Planck Institute (MPI) for Biochemistry she became Group Leader at the MPI for Biology of Ageing in 2010. In 2018 her laboratory moved to the University of Helsinki where she was professor of Cell and Developmental Biology. In 2022 Wickström was appointed as Director of the MPI for Molecular Biomedicine in Münster. Research in the Wickström lab aims to understand how mammalian epithelial tissues are generated and maintained. We particularly focus on how mechanical forces and cellular interactions integrate single cell behaviours to pattern these structurally extremely robust yet dynamic tissues. |
| 09:25-09:40 |
Discussion
|
| 09:40-10:00 |
Enabling neighbour labelling: leveraging synthetic biology to monitor and manipulate cell-cell interactions
Cell-cell interactions profoundly influence cellular decision-making during development, in homeostasis, and during the onset and progression of disease. However, identifying these interactions and decoding their downstream effects remains technically challenging due to a shortage of relevant analytical approaches. We have developed a series of engineering biology tools that make it possible to fluorescently label cells that neighbour any cell of interest. This allows us, for example, to fluorescently label healthy cells interacting with mutant cells, or pluripotent cells interacting with differentiated cells. Live labelled neighbours and unlabelled non-neighbours can be isolated by flow cytometry for further downstream analysis; this includes both ‘omics studies and functional assays on these live interacting cells. The establishment of this type of technology relies on finely tuned genetic circuits, and requires iterative comparison of different transgenic structures through a series of design-build-test-learn cycles. Historically, these cycles have been complicated by “apples to pears” comparisons of different transgenes delivered episomally or randomly integrated in various genomic regions, which confound results due to variable delivery efficiencies, differing levels of transgene expression, and transgene silencing. To overcome this hurdle, we generated clonal “master” cell lines harbouring two insulated modular genomic landing pads integrated in safe harbour sites. These lines allow for rapid and convenient transgene integration in the same genomic loci, removing all variability caused by episomal delivery or random integration, and allowing for true "apples to apples" comparisons. 
Dr Matthias MalagutiUniversity of Edinburgh, UK 
Dr Matthias MalagutiUniversity of Edinburgh, UK Mattias Malaguti has spent his entire career at the University of Edinburgh. From undergraduate research in Tom Burdon’s lab (Roslin Institute) through PhD and postdoctoral studies in Sally Lowell’s lab (Institute for Stem Cell Research), he has consistently focused on developing and adapting novel technologies for use in developmental biology models, aiming to address previously intractable biological questions. Driven by an interest in how neighbouring cells influence each other's decisions, and frustrated by quantitative imaging limitations in growing embryos, Mattias recently developed engineering biology tools to identify and isolate live interacting cells. He now leads his own group at the Centre for Engineering Biology, where he leverages these innovative tools to investigate how healthy cells sense and respond to mutant neighbours in models of development and disease. |
| 10:00-10:15 |
Discussion
|
| 10:15-10:40 |
Break
|
| 10:40-11:00 |
Reconstituting tissue-scale signalling landscapes from the bottom up
Tissues are not random assemblies of cells; instead, they are highly organized structures in which cells of distinct types are spatially patterned to create tissue form and function. This organization is governed by precise, dynamic patterns of signalling activity that unfold across space and time. For simplicity, I refer to these coordinated signalling activities as a tissue’s signalling landscape. These landscapes shape how cells acquire their identities, interact with one another, and ultimately give rise to tissue architecture and function. While the core genetic components of key developmental signalling pathways are well studied, it remains unclear how they act together to establish the signalling landscapes. For example, how are such landscapes created with spatial precision and adapted for diverse tissue contexts? To address these questions, my lab uses synthetic reconstitution approaches to build signalling landscapes from the ground up in genetically minimal, “blank-slate” cells. By reconstructing modules, such as morphogen gradients and planar cell polarity systems, we aim to identify the minimal set of components required for the formation of signalling landscapes and uncover the general principles that govern their dynamics and evolution. In this talk, I will present our recent findings and the broader implications for understanding tissue patterning. 
Dr Pulin LiWhitehead Institute, MIT, US 
Dr Pulin LiWhitehead Institute, MIT, US Pulin Li is the Eugene Bell Career Development Professor of Tissue Engineering at MIT and a member of the Whitehead Institute. She completed a PhD in Chemical Biology from Harvard University and did her postdoctoral work at Caltech. Her lab studies how cell-cell communication spatially organizes cells into patterns and shapes, by reconstituting such phenomena from the bottom up. Using quantitative imaging, synthetic engineering, and mathematical modelling, her lab asks two major questions: (1) How to build signalling pathways with self-organizing capabilities using molecular parts? (2) How to build structures of defined shapes and functions by putting different types of cells together, such as the branched structure of the lung? Her research program aims to provide fundamental insights into the principles of embryo development, and the tools necessary for engineering more sophisticated self-organization of organ models in a dish. |
| 11:00-11:15 |
Discussion
|
| 11:15-12:00 |
Invited abstracts
|
| 13:40-14:00 |
Engineering gastrulation: Exploring the limits of morphogenetic robustness to altered embryo geometry
Changes in reproductive strategy are common across the evolution of vertebrate development. How this impacts the morphogenetic processes of gastrulation in manner that enables the emergence of a common body plan are not well understood. Gastruloids reduce the morphological complexity of pre-gastrulation embryos, can be compared across species and allow for a systematic exploration of the impact of changes in tissue geometry, ECM and mechanical environment. This enables us to determine the degree to which tissue-level flows associated with gastrulation are robust to the experimental disruption of these boundary conditions, and those that emerge through the interactions of cells with the acellular niche. To extract information about tissue-level flows, we have developed tools to conduct three-dimensional particle velocimetry analysis and applied them to zebrafish blastoderm explants (pescoids). We observed polonaise-like symmetric flows that become interrupted at the onset of elongation. Computational modelling indicates that converging and internalising flows are active forces that synergise to break these symmetric flows and sustain elongation. Consistently, we observe cell movements associated with both convergence and internalisation during explant elongation. I will also present work in mouse gastruloids that explores the role of substrate stiffness and ECM composition on the self-organisation of collective cell migration. We isolate conditions that enable the self-generation of multiple protrusions, that each self-organise a tailbud-like morphology with associated morphogen gradient systems. Together, our work determines examples of both robust genetically encoded morphogenesis, and the self-organisation of well conserved patterning mechanisms from altered starting conditions. 
Dr Ben SteventonUniversity of Cambridge, UK 
Dr Ben SteventonUniversity of Cambridge, UK Ben Steventon trained in the labs of Roberto Mayor (as PhD 2004-2008, UCL, UK), Andrea Streit (as PostDoc 2008-2012, KCL, UK), Jean-Francois Nicolas and Estelle Hirsinger (as PostDoc 2011-2013, Institute Pasteur, France), Scott Fraser (as Marie-Curie Outgoing Fellow 2013-2014, USC, USA) and Alfonso Martinez Arias (as Marie-Curie Incoming Fellow 2014-2015, Cambridge). He started his research group in 2016 supported by a Wellcome Trust/Royal Society Sir Henry Dale fellowship. In November 2021 he transitioned to an Assistant Professor in the Department of Genetics (University of Cambridge). |
|---|---|
| 14:00-14:15 |
Discussion
|
| 14:15-14:35 |
Towards in vitro reconstitution of Wnt gradient formation
Wnt proteins are essential regulators of organ development, homeostasis, and repair. They feature a unique post-translational lipid modification (palmitoleoylation), essential for engaging their signaling receptor Frizzled and consequently downstream signal transduction. Despite the inherent hydrophobicity caused by this lipid moiety, Wnts can spread in the extracellular space and act over many cell diameters in different tissue contexts. Work from our lab has shown that the glypican Dlp harbours a hydrophobic tunnel that stabilizes Wingless (Wg), the main Drosophila Wnt protein, at the cell surface and thus contributes to signalling activity. We found that glycosaminoglycans (GAG) chains can ‘solubilise’ Wingless independently of the lipid-binding tunnel of Dlp and that, in gain-of-function experiments, glypicans GAGs promote the spreads of Wingless along the surface of Drosophila wing imaginal discs and drive extracellular gradient formation. We are interested in uncovering the molecular mechanism that enable GAGs alone to promote Wg transport. Biophysical analysis of the GAG-Wg interaction suggests that GAGs induce the formation of multimeric Wg complexes, a conformation that may allow reciprocal lipid-shielding by Wg molecules. Our ultimate goal is to combine genetic engineering approaches, mathematical modelling, and in vitro reconstitution assays to reach a mechanistic understanding of how lipid- and GAG-dependent interactions cooperate to enable the extracellular transport of Wingless, and other Wnts. 
Dr Jean-Paul Vincent FMedSci FRSThe Francis Crick Institute, UK 
Dr Jean-Paul Vincent FMedSci FRSThe Francis Crick Institute, UK After completing an M.Eng. in applied physics at the University of Louvain (Belgium), Jean-Paul obtained a Fulbright fellowship for postgraduate studies in Biophysics at U.C. Berkeley where he characterized the symmetry-breaking events that initiate frog development. As a post-doctoral Damon Runyon fellow with Patrick O’Farrell at UC San Francisco, he devised a caged dye approach to label single cells and study cell fate specification. Since moving to the UK as a group leader, first at MRC-LMB, then at MRC-NIMR, and currently at the Francis Crick Institute, Jean-Paul has been studying how cells signal to each other to ensure correct patterning and growth during development. He is interested in the biophysics of morphogen gradient formation, with a particular on a class of signaling proteins called Wnts. He also studies how signals involved in normal development mediate the response to tissue stress. Jean-Paul is a fellow of the Royal Society and the Academy of Medical Sciences. He co-founded Summit Therapeutics in 2001. |
| 14:35-14:50 |
Discussion
|
| 14:50-15:10 |
Break
|
| 15:10-15:30 |
Synthetic gene regulatory networks to explore design principles of spatiotemporal patterning
Patterns are ubiquitous in biology ranging from gene expression during development to the organization of microbial communities. While the molecular mechanisms underlying pattern formation are complex, they are believed to follow general principles. In my group we use a bottom-up synthetic biology approach to construct in bacteria synthetic gene regulatory networks capable of generating spatiotemporal patterns. We have for example worked on stripe-forming incoherent feed-forward loops, on bistable switches and on oscillators. This strategy not only enables precise control over pattern formation but also provides insights into the fundamental principles that govern these biological processes. In my talk, I will give an overview of some of our recent work on pattern formation. 
Professor Yolanda SchaerliUniversity of Lausanne, Switzerland 
Professor Yolanda SchaerliUniversity of Lausanne, Switzerland Professor Yolanda Schaerli is Associate Professor at the University of Lausanne, Switzerland in the Department of Fundamental Microbiology, where she heads a research group in bacterial synthetic biology. Professor Schaerli studied at the ETH Zurich, Switzerland and at Imperial College, London, UK before she carried out a PhD at the University of Cambridge, UK. She then joined the Centre for Genomic Regulation (CRG), Barcelona, Spain, to do a postdoc. In 2014 she returned to her home country as junior group leader at the University of Zurich. In 2017 she started as Tenure-Track Assistant Professor at the University of Lausanne and was promoted to Associate Professor in 2023. Research in her laboratory uses synthetic biology to engineer synthetic circuits in bacteria, with a focus on gene regulatory networks involved in spatiotemporal pattern formation. The group also develops novel tools and approaches for synthetic biology. |
|---|---|
| 15:30-15:45 |
Discussion
|
| 15:45-16:05 |
Information flow in self-organized developmental systems
Embryonic development is a spectacular display of self-organisation of multi-cellular systems, combining transformations of tissue mechanics and patterns of gene expression. These processes are driven by the ability of cells to communicate through mechanical and chemical signalling, allowing coordination of both collective movement and patterning of cellular states. To ensure proper biological function, such patterns must be established reproducibly, by controlling and even harnessing intrinsic and extrinsic fluctuations. While the relevant molecular processes are increasingly well understood, we lack principled frameworks to understand how tissues obtain information to generate reproducible patterns. I will discuss how combining dynamical systems models with information theory provides a mathematical language to analyse biological self-organization across diverse systems. Our approach can be used to define and measure the information content of observed patterns, to functionally assess the importance of various patterning mechanisms, and to predict optimal operating regimes of self-organizing systems. I will demonstrate how our framework reveals mechanisms of self-organization of in vitro stem cell systems in direct connection to experimental data, including intestinal organoids and gastruloids. This framework provides an avenue towards unifying the zoo of chemical and mechanical signalling processes that orchestrate embryonic development. 
Professor David BrücknerBiozentrum, University of Basel, Switzerland 
Professor David BrücknerBiozentrum, University of Basel, Switzerland David Brückner is an Assistant Professor of Theoretical Biophysics at the Biozentrum and Department of Physics of the University of Basel, Switzerland. Previously, he was an EMBO and NOMIS Foundation Postdoctoral Fellow at the Institute of Science and Technology Austria. He performed his doctoral studies at the Ludwig Maximilian University in Munich. He has been awarded with the Gustav-Hertz-Prize of the German Physical Society. |
| 16:05-16:20 |
Discussion
|
| 16:20-16:40 |
Synthetic biology tools for mammalian cell engineering
Our research at Imperial College is focused on the engineering of bacteria, mammalian and synthetic cells to advance the applications of synthetic biology. Resource competition between host cells and genetic constructs is one of our main research themes, for its impact on our ability to reliably engineer cellular hosts. In order to tackle resource competition and the cellular burden derived from it, we are currently developing new tools to address measure and characterise competition in engineered cells, but also exploring the design of novel gene expression control platforms for low-footprint genetic engineering. 
Dr Francesca CeroniImperial College London, UK 
Dr Francesca CeroniImperial College London, UK Francesca graduated in Pharmaceutical Biotechnology and received a PhD in Bioengineering from the University of Bologna before moving to Imperial College London for her PDRA. She is currently a Senior Lecturer in the Department of Chemical Engineering at Imperial College London. She has track record in host-aware gene construct design for synthetic biology. Her group currently focuses on developing novel strategies for improved gene expression control in bacteria and mammalian. |
| 16:40-16:55 |
Discussion
|
| 16:55-17:15 |
Flash poster talks
|
|---|---|
| 17:15-18:15 |
Poster session
|
Chair

Professor Sally Lowell
University of Edinburgh, UK

Professor Sally Lowell
University of Edinburgh, UK
Sally Lowell is a Wellcome Trust Senior Fellow and Professor of Stem Cell Biology and Early Development, based in the Centre for Regenerative Medicine. She did her PhD with Fiona Watt at CRUK followed by postdoctoral positions with David Anderson at Caltech and Austin Smith in Edinburgh. She is a Director at the Company of Biologists, where she chairs their Sustainability Initiative. She is also an elected member of EMBO and a Fellow of the Academy of Medical Sciences. Her lab build models based on pluripotent cells in order to investigate how cells coordinate with their neighbours to build tissues during development, and how cells sense and respond to pathological changes in neighbours in disease.
| 09:05-09:25 |
From molecules to morphogenesis: Decoupling the principles of early mammalian development
Classical models of embryonic patterning emphasize transcription factors and morphogen gradients as primary drivers of spatial and temporal gene expression. Yet, emerging evidence suggests that metabolism plays a more instructive role than previously appreciated. We recently uncovered two distinct, stage- and cell-type-specific waves of glucose metabolism that orchestrate key developmental transitions during mouse gastrulation. Using quantitative single-cell imaging, stem cell models, and tissue explants, we show that hexosamine pathway activity in the epiblast promotes fate acquisition, while a later glycolytic wave supports mesoderm migration and tissue expansion. These metabolic programs regulate development through distinct cellular signalling mechanisms. Our findings position compartmentalized metabolism as a dynamic and instructive layer in embryonic patterning, acting in concert with genetic and signalling cues to shape the mammalian body plan. 
Dr Berna SozenYale University, US 
Dr Berna SozenYale University, US Berna Sozen is an Assistant Professor at Yale University Departments of Genetics and Reproductive Sciences. She earned her BSc in Biology and MSc in Reproductive Biology at Akdeniz University in Turkey. In 2015, she joined the University of Cambridge to conduct her PhD thesis focused on early embryogenesis and stem cell biology, under the mentorship of Professor Magdalena Zernicka‑Goetz. After completing her PhD and postdoctoral work—with stints at Cambridge and Caltech—she joined Yale in 2020. Her research group studies early mammalian development, metabolism, and maternal–foetal interactions using mouse and human embryos, as well as stem cell-derived embryo-like models. |
|---|---|
| 09:25-09:40 |
Discussion
|
| 09:40-10:00 |
Towards light-guided physiology: optogenetics and biosensors to probe tissue-scale patterning
How do cells organise into distinct spatial domains in a healing wound or developing embryo? We seek to understand how the local activation of signalling pathways influences cell decisions and “re-write” this information to drive desired cellular responses. Both missions require new tools: biosensors to report on cells’ states, and optogenetic or chemical tools to alter them. I will talk about our recent work on all three areas – optogenetic tools, biosensors, and cell signalling biology – with a particular focus on receptor tyrosine kinases in development. 
Professor Jared ToettcherPrinceton University, US 
Professor Jared ToettcherPrinceton University, US Jared Toettcher is an Associate Professor of Molecular Biology and Deputy Director of the Omenn-Darling Bioengineering Institute at Princeton University. He obtained a PhD from MIT working with Bruce Tidor (MIT) and Galit Lahav (Harvard Medical School), and was a postdoctoral fellow at UCSF with Wendell Lim and Orion Weiner. He has been named a 2016 NIH New Innovator, a 2019 Vallee Scholars, and a 2024 Allen Distinguished Investigator. |
| 10:00-10:15 |
Discussion
|
| 10:15-10:40 |
Break
|
| 10:40-11:00 |
Talk title TBC
|
| 11:00-11:15 |
Discussion
|
| 11:15-12:00 |
Invited abstracts
|
| 11:35-11:50 |
Discussion
|
| 13:40-14:00 |
Designing purpose and agency in engineered generative biology
A characteristic of living systems is that they act with internal purposive behaviour and agency. Cells, cell collectives and organisms have agendas; using a combination of environmental sensing and internal parameters, including parameters set by part events and persisting, they navigate through state space toward goals. Natural generative biological systems such as embryos demonstrate this in their responses to experimental perturbation; when moved from their natural trajectory through state-space, they generally adapt to find a new trajectory to their original goal (Waddington called this 'homeorhesis'). Designers of engineered biological systems can choose where to locate purpose and agency for the biological device that they build, They can keep it outside biology, for example by using manual approaches or closed-loop, computer-driven systems to control engineered cells with chemical signals or with light. Or – and this is critical for truly generative engineered biological systems – they can include goal-seeking/ agential behaviour as part of their biological design, often (at this stage of technology) as an adaptation of existing natural agential systems. In this talk, I will briefly review concepts of purpose and agency, and then discuss how they can be handled in the design of generative biological systems. The principles will be illustrated with real examples, constructed in wet-ware by my own and other laboratories. 
Professor Jamie Davies FRSEUniversity of Edinburgh, UK 
Professor Jamie Davies FRSEUniversity of Edinburgh, UK Jamie Davies is Professor of Experimental Anatomy at the University of Edinburgh. He is a scientist-engineer with an interest in how simple things become complicated. His lab studies organ development, by ‘wet-lab’ techniques and computer modelling, and applies knowledge gained to the problem of engineering realistic tissues and organs from stem cells. This is coupled to an interest in engineering completely novel forms of development using the techniques of synthetic biology. The lab also hosts the main drug database of the International Union of Basic and Clinical Pharmacology. He has published over 200 research papers and 11 books on these topics. |
|---|---|
| 14:00-14:15 |
Discussion
|
| 14:15-14:45 |
Invited abstracts
|
| 14:45-15:00 |
Break
|
| 15:00-15:20 |
Evolving phenotypic diversity
One of the fundamental and most longstanding questions in biology is how phenotypes evolve to generate the incredible diversity of forms that surround us in the natural world. Vertebrate body shape is a good example of this, with species’ forms ranging from the rounded ocean sunfish to elongated snakes, and everything in between. The evolution of vertebrate body shapes has been accompanied by significant variation to the axial skeleton and the vertebrae that compose it, which can vary in size, morphology and type, but crucially also in number, ranging from less than ten to several hundred. In my group we take an interdisciplinary approach to uncover the developmental drivers of phenotypic diversity in vertebrates using Lake Malawi cichlid fishes as a model system to investigate the evolution of increased vertebral counts. The diverse morphology, conserved genomes, close-relatedness, and ability to hybridise of these fishes makes them the ideal system in which to study how the evolutionary modification of developmental mechanisms has led to phenotypic diversity, in this case by evolving increased vertebral counts. We combine state-of-the-art experimental embryology, single cell technologies, advanced microscopy, ancestral trait reconstruction and data-driven mathematical modelling to ask how developmental processes differ in Lake Malawi cichlid species that have convergently evolved increased vertebral counts, versus those that have retained ancestral numbers of vertebrae. Specifically, we focus on two developmental processes known to play important roles determining total vertebral counts - gastrulation and somitogenesis - and use data-driven mathematical modelling to ask how these differences might have evolved. The interdisciplinary projects carried out in my group are already contributing towards our understanding of how phenotypic diversity is generated and have the potential to inform how phenotypes might evolve in the future. 
Dr Berta VerdUniversity of Oxford, UK 
Dr Berta VerdUniversity of Oxford, UK Dr Verd is a somewhat unusual breed of biologist. A mathematician by training, she spent a year studying a Masters degree in sociology of science before moving into biology during her second Masters degree in Systems and Synthetic Biology at Imperial College, London. Through her Masters research, it became clear to her that interdisciplinary approaches held huge potential to help us understand the central problems in biology, and she's been hooked ever since. She moved to the Centre for Genomic Regulation (CRG) in Barcelona to pursue a doctorate degree in evolutionary and developmental systems biology under the supervision of Dr Johannes Jaeger. During her PhD she used data-driven mathematical modeling to study pattern formation during segment determination in flies. She developed mathematical tools to characterise gene expression dynamics, allowing them to compare these amongst different arthropod species. This work helped them understand how gene regulatory networks drive gene expression dynamics in developmental processes and shape their evolution. In October 2017 she joined the Steventon Lab in the Department of Genetics at the University of Cambridge as a Herchel-Smith Postdoctoral Fellow where she combined experimental embryology, microscopy and dynamical modelling to understand axial elongation and patterning in zebrafish and cichlid embryos. Since April 2020, she is the Associate Professor of Computational and Theoretical Biology in the Department of Zoology at the University of Oxford, and a Tutorial Fellow in the Biological Sciences at Jesus College. |
| 15:20-15:35 |
Discussion
|
| 15:35-15:55 |
Talk title TBC
|
| 15:55-16:10 |
Discussion
|
| 16:10-17:30 |
Round table discussion: An agenda for generative biology

Dr Philip BallFreelance writer 
Dr Philip BallFreelance writer Philip Ball is a freelance writer and broadcaster, and was an editor at Nature for more than twenty years. He writes regularly in the scientific and popular media and has written many books on the interactions of the sciences, the arts, and wider culture, including H2O: A Biography of Water, Bright Earth: The Invention of Colour, The Music Instinct, and How Life Works. His book Critical Mass won the 2005 Aventis Prize for Science Books. Ball was the 2022 recipient of the Royal Society’s Wilkins-Bernal-Medawar Medal for contributions to the history, philosophy or social roles of science. He trained as a chemist at the University of Oxford and as a physicist at the University of Bristol. He lives in London. |





